amino acid to dna sequence converter
Peptide Amino Acids Sequence Converter. Ad 5000x more data than other providers.
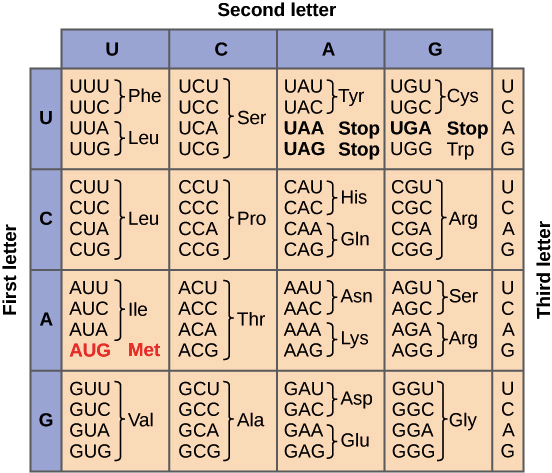
Overview Of Translation Article Khan Academy
Apologies for the inconvenience.
. Coding strand or non-coding strand. If you have only a DNA sequence to find the corresponding amino acids first transcribe your DNA sequence into an RNA sequence using complimentary base pairing. A consensus sequence derived from all the possible codons for each amino acid is also returned.
Macromoltek - Revolutionizing antibody design. Convert a DNA sequence into Amino Acid. Few steps to find amino acid sequence.
Macromoltek started in 2010 and has since built and refined antibody design and modeling software. For amino acids with multiple possible nucleotide. I am then to find the amino acid that these DNA sequences encode per codon each group of three literals For example if I have a.
Note that amino acids with 6 synonymous codons Leu Arg and Ser cannot be represented by a single unambiguous degenerate sequence. DNA d eoxyribo n ucleic a cid permanent copy of the genetic information. Reverse task may be solved with a program Six-Frame Translation.
This will allow you to convert a GenBank flatfile gbk to GFF General Feature Format table CDS coding sequences Proteins FASTA Amino Acids faa DNA sequence Fasta format. Translates DNA or mRNA to the other and a Protein strand amino acids. Galaxy is an open web-based platform for accessible reproducible and transparent computational biomedical research.
Toward this we demonstrate successful target identification with two sets of target-binder pairs. I added this feature because the In general the mapping from an amino acid to a nucleotide codon is not a one- to-one mapping. Viewed 828 times -3 I am trying to write a bash script that would be able to read DNA sequences each line in the file is a sequence from a file where sequences are separated by an empty line.
This program examines the input sequence in all six possible frames ie. One to Three converts single letter translations to three letter. Back-translation is used to predict the possible nucleic acid sequence that a specified peptide sequence has originated from.
Ask Question Asked 6 years 4 months ago. Galaxy is an open web-based platform for accessible reproducible and transparent computational biomedical research. Also I came across How do I find the nucleotide sequence of a protein using Biopython but this is what I am not looking for.
For example Leu can be encoded by CTN or. Due to scheduled maintenance work this service will not be available on Monday February 7 between 0745 am and 0930 am CEST. Three to One converts three letter translations to single letter translations.
Each three base sequence of the mRNA called a codon is read by the ribosome and the appropriate amino acid is inserted into the growing protein. DNA to mRNA to Protein Converter. Sequence Translation is used to translate nucleic acid sequence to corresponding peptide sequences.
A sequence can represent either a nucleic acid eg DNA or RNA or a polypeptide chain representing amino acids. I am trying to write a bash script that would be able to read DNA sequences each line in the file is a sequence from a file where sequences are separated by an empty line. The Amino Acid Converter can be used to convert sequences of amino acid residues between 1 and 3 letter notation.
A Google search gives me result of DNA to protein conversion but not vice versa. Translate is a tool which allows the translation of a nucleotide DNARNA sequence to a protein sequence. DNA and RNA codon to amino acid converter.
Convert a DNA sequence into Amino Acid. Reverse translation of aminoacid sequences This program helps to predict nucleic acid sequence for the given amino acid sequence. Digits and blank spaces are removed automatically.
DNA binders deposit unique amino acid-identifying barcodes on the chip. At the upload sequences page select the Amino Acid Sequence option. You can create an empty list to keep the results for the translation of each codon eg.
Our website will accept amino acid sequences and convert them to a DNA sequence for synthesis. Three-Letter to One-Letter One-Letter to Three-Letter Purpose. This will allow you to convert a GenBank flatfile gbk to GFF General Feature Format table CDS coding sequences Proteins FASTA Amino Acids faa DNA sequence Fasta format.
We support only the standard 20 one-letter code characters. I want to convert a list of fasta protein sequences in a text file into corresponding nucleotide sequences. STEP 1 Know which DNA strand is given.
Nucleotide Sequence Translation Transeq EMBOSS EMBOSS Transeq translates nucleic acid sequences to the corresponding peptide. Converts sequences from DNA to RNA and from this to protein. STEP 2 Write the corresponding m-RNA strand.
You got the amino acid output for the first codon only because you used return inside the for loop. Reading the sequence from 5 to 3 and from 3 to 5 starting with nt 1 nt 2 and nt 3. For manipulations with nucleic acid sequence reverse reversecomplement double stranded it is possible to use Sequence Utilities program.
2 Degenerate codons. Help with amino acid. Make sure your input only contains Amino Acid one-letter characters.
I am then to find the amino acid that. Our team of engineers and scientists have a plethora of knowledge in biology and computational sciences. Non-standard triplets are ignored.
The end goal is that over multiple binding cycles a sequential chain of DNA barcodes will identify the amino acid sequence of a peptide. Once the first amino acid is returned the loop terminates hence the second codon wont be tested at all. Reverse Translate accepts a protein sequence as input and uses a codon usage table to generate a DNA sequence representing the most likely non-degenerate coding sequence.
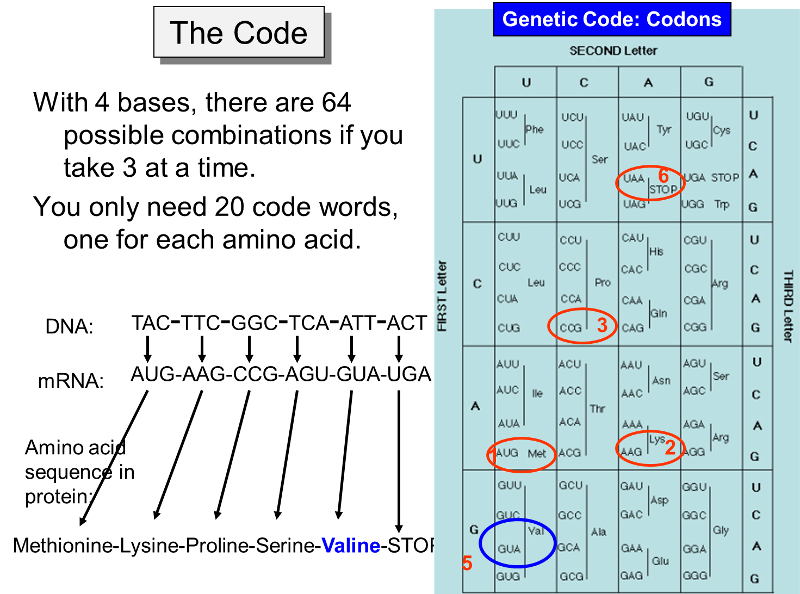
Active 6 years 4 months ago. There are two strands. The central dogma of biology states that a segment DNA is transcribed and translated into a protein according to the following code in which every three letters of DNA except three cases become one letter of amino acid.
This method will create a degenerate sequence and where possible each codon will encode all possible synonymous codons for a particular amino acid. Translating the DNA sequence is done by reading the nucleotide sequence three bases at a time and then looking at a table of the genetic code to arrive at an amino acid sequence. One can either read the coding strand from 3 to 5 or read the template strand from 5 to 3 when making the corresponding m-RNA strand.
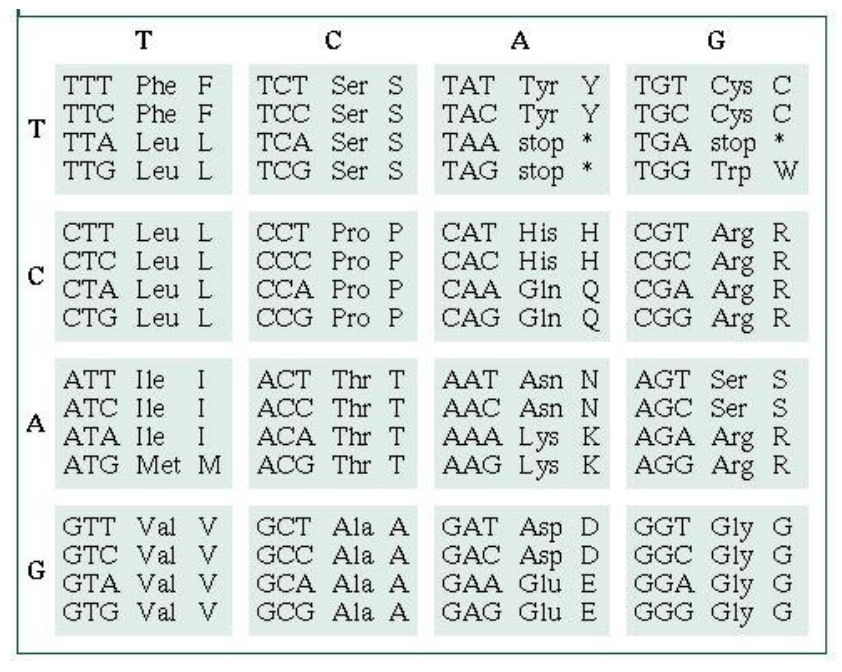
Genetics Deducing Amino Acid Sequence From A Dna Sequence Biology Stack Exchange

Decode From Dna To Mrna To Trna To Amino Acids Youtube

How Can Amino Acid Sequence Be Determined From Dna Quora

The Dna Sequence Of A Gene Encodes The Amino Acid Sequence Of A Protein Download Scientific Diagram
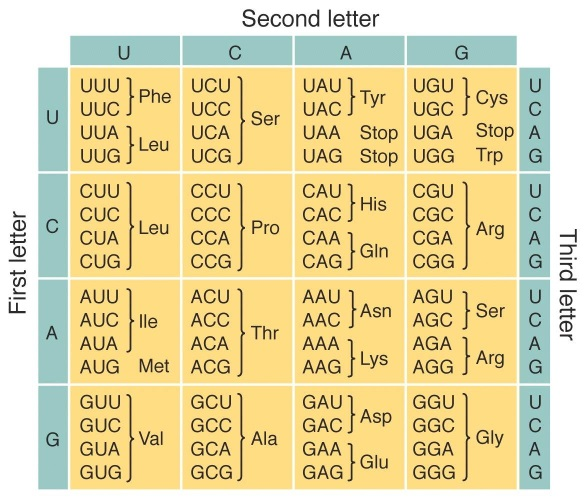
Solved Dna Transcription And Translation Directions 1 Chegg Com
What Would The Amino Acid Sequence Be Specified By The Transcribed Dna Sequence Quora

Genomics And Comparative Genomics
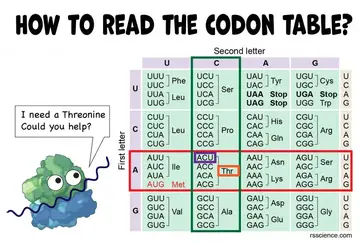
How To Read The Amino Acids Codon Chart Genetic Code And Mrna Translation Rs Science
3 5 Transcription And Translation Bioninja
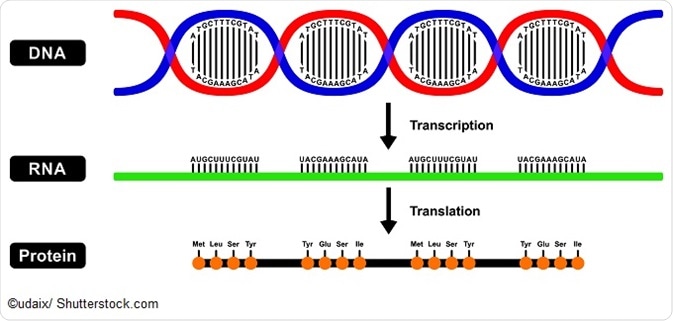
Amino Acids And Protein Sequences

How To Get Dna Sequence From Protein Sequence Quora

Science Olympiad Protein Modeling Event The Molecular Story Of Xiap
How To Get Dna Sequence From Protein Sequence Quora
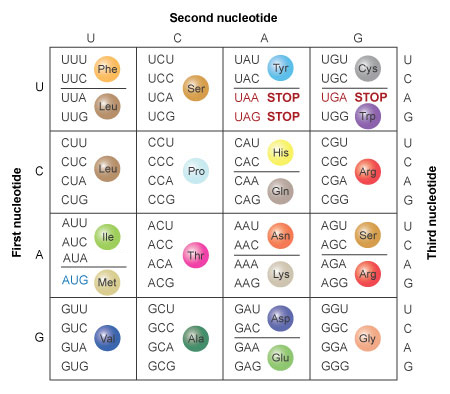
Nucleic Acids To Amino Acids Dna Specifies Protein Learn Science At Scitable

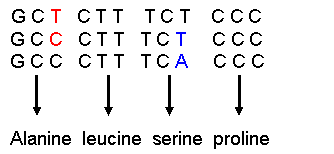

Comments
Post a Comment